
odo Documentation
Release 0.5.0+26.g55cec3c
Matthew Rocklin
August 11, 2016

ii
odo Documentation, Release 0.5.0+26.g55cec3c
odo takes two arguments, a source and a target for a data transfer.
>>> from odo import odo
>>> odo(source, target) # load source into target
It efficiently migrates data from the source to the target through a network of conversions.
Contents 1

odo Documentation, Release 0.5.0+26.g55cec3c
2 Contents

CHAPTER 1
Example
>>> from odo import odo
>>> import pandas as pd
>>> odo('accounts.csv', pd.DataFrame) # Load csv file into DataFrame
name balance
0 Alice 100
1 Bob 200
2 Charlie 300
>>> # Load CSV file into Hive database
>>> odo('accounts.csv', 'hive://user:password@hostname/db::accounts')
3

odo Documentation, Release 0.5.0+26.g55cec3c
4 Chapter 1. Example
CHAPTER 2
Contents
2.1 General
2.1.1 Project Information
Getting Odo
• Releases
$ conda install odo
• Development Packages
$ conda install odo -c blaze
• PIP
$ pip install odo
$ pip install git+git://github.com/blaze/odo
Source
Odo development takes place on GitHub: https://github.com/blaze/odo.
Reporting Issues
• Bugs and feature requests can be filed here.
• The blaze development mailing list is good place to discuss ideas and ask questions about odo.
2.1.2 Overview
Odo migrates between many formats. These include in-memory structures like list, pd.DataFrame and
np.ndarray and also data outside of Python like CSV/JSON/HDF5 files, SQL databases, data on remote machines,
and the Hadoop File System.
5
odo Documentation, Release 0.5.0+26.g55cec3c
The odo function
odo takes two arguments, a source and a target for a data transfer.
>>> from odo import odo
>>> odo(source, target) # load source into target
It efficiently migrates data from the source to the target.
The target and source can take on the following forms
Source Target Example
Object Object An instance of a DataFrame or list
String String
Type
’file.csv’, ’postgresql://hostname::tablename’ list,
DataFrame
So the following lines would be valid inputs to odo
>>> odo(df, list) # create new list from Pandas DataFrame
>>> odo(df, []) # append onto existing list
>>> odo(df, 'myfile.json') # Dump dataframe to line-delimited JSON
>>> odo('myfiles.
*
.csv', Iterator) # Stream through many CSV files
>>> odo(df, 'postgresql://hostname::tablename') # Migrate dataframe to Postgres
>>> odo('myfile.
*
.csv', 'postgresql://hostname::tablename') # Load CSVs to Postgres
>>> odo('postgresql://hostname::tablename', 'myfile.json') # Dump Postgres to JSON
>>> odo('mongodb://hostname/db::collection', pd.DataFrame) # Dump Mongo to DataFrame
Warning: If the target in odo(source, target) already exists, it must be of a type that supports in-place
append.
>>> odo('myfile.csv', df) # this will raise TypeError because DataFrame is not appendable
Network Effects
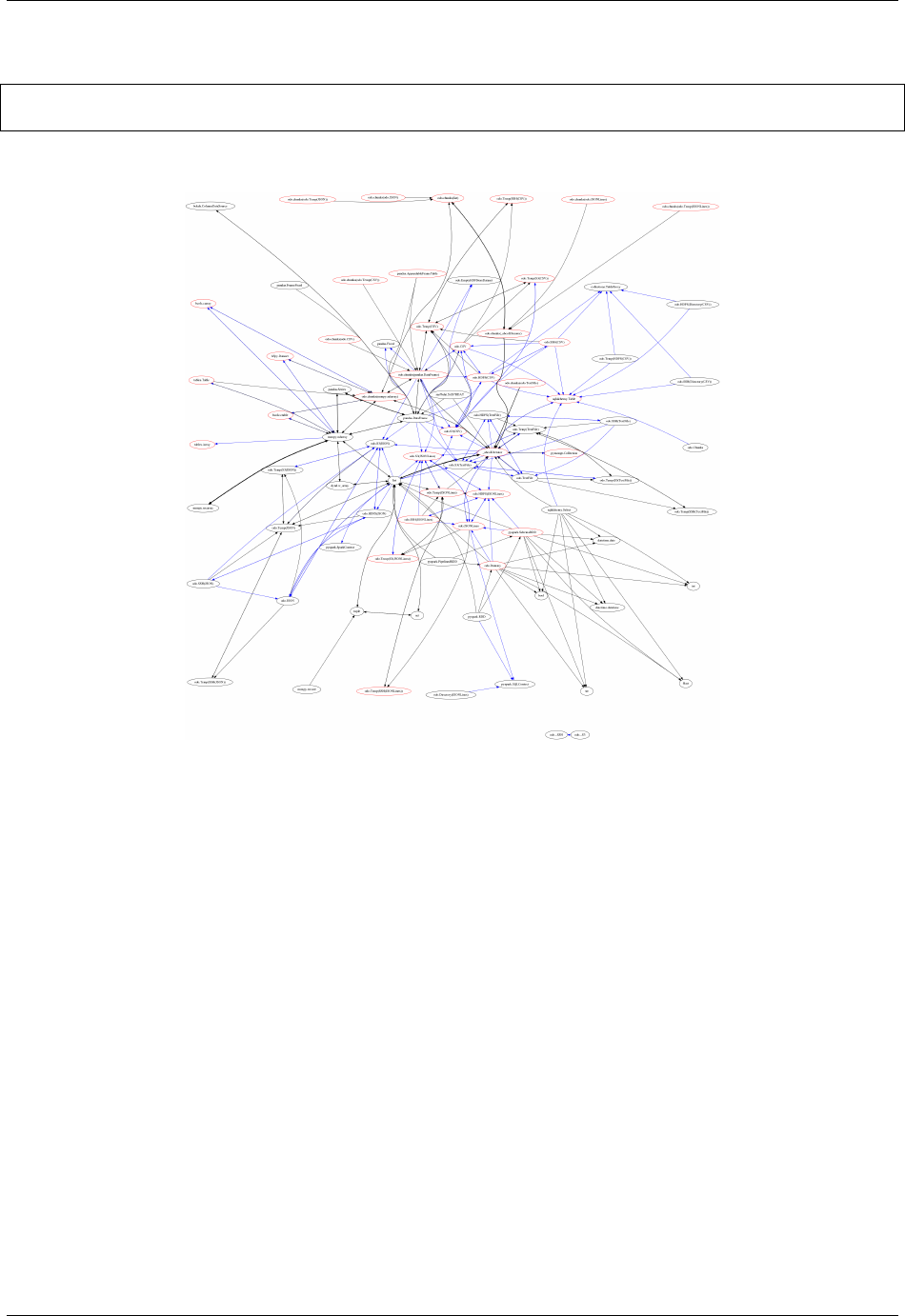
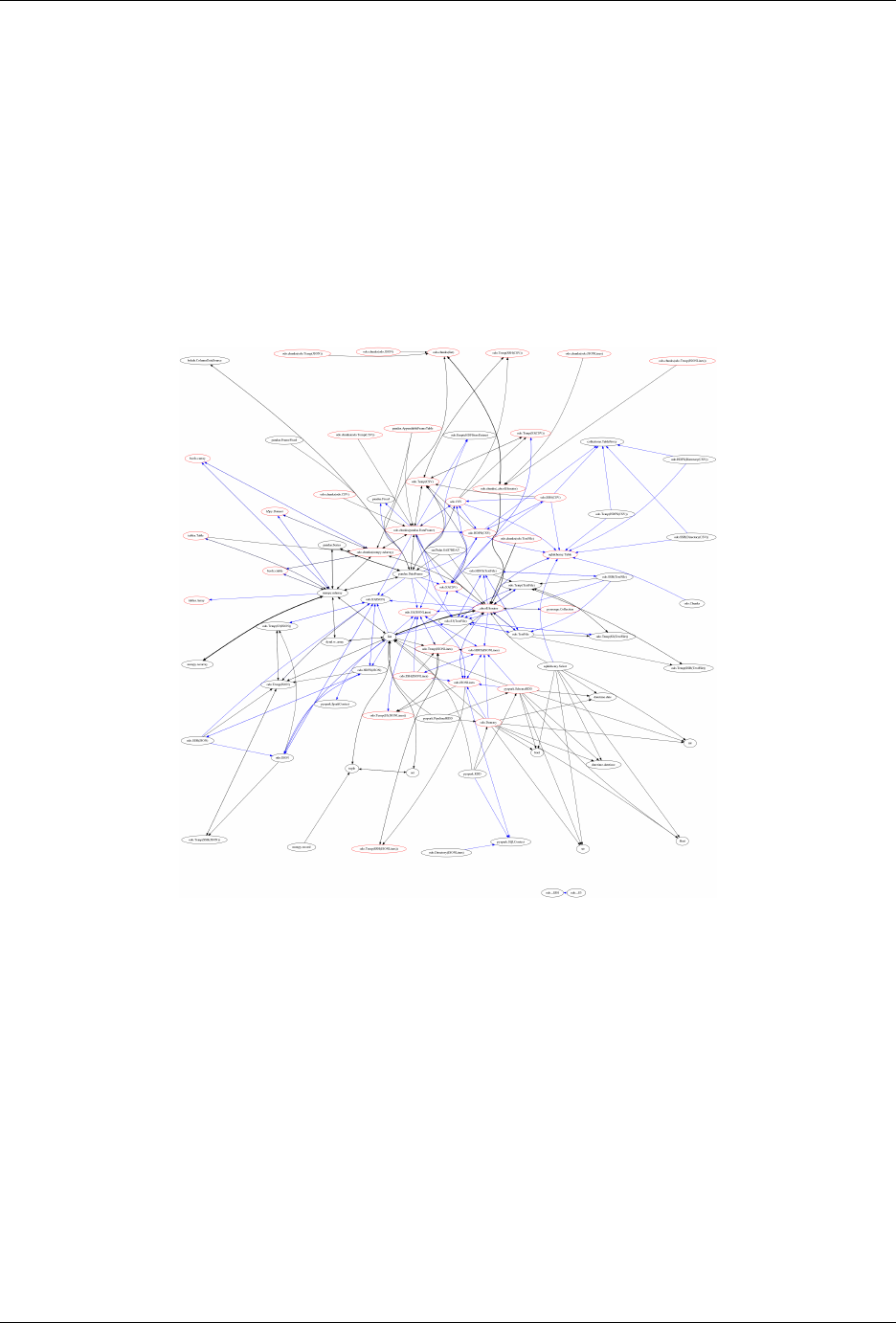
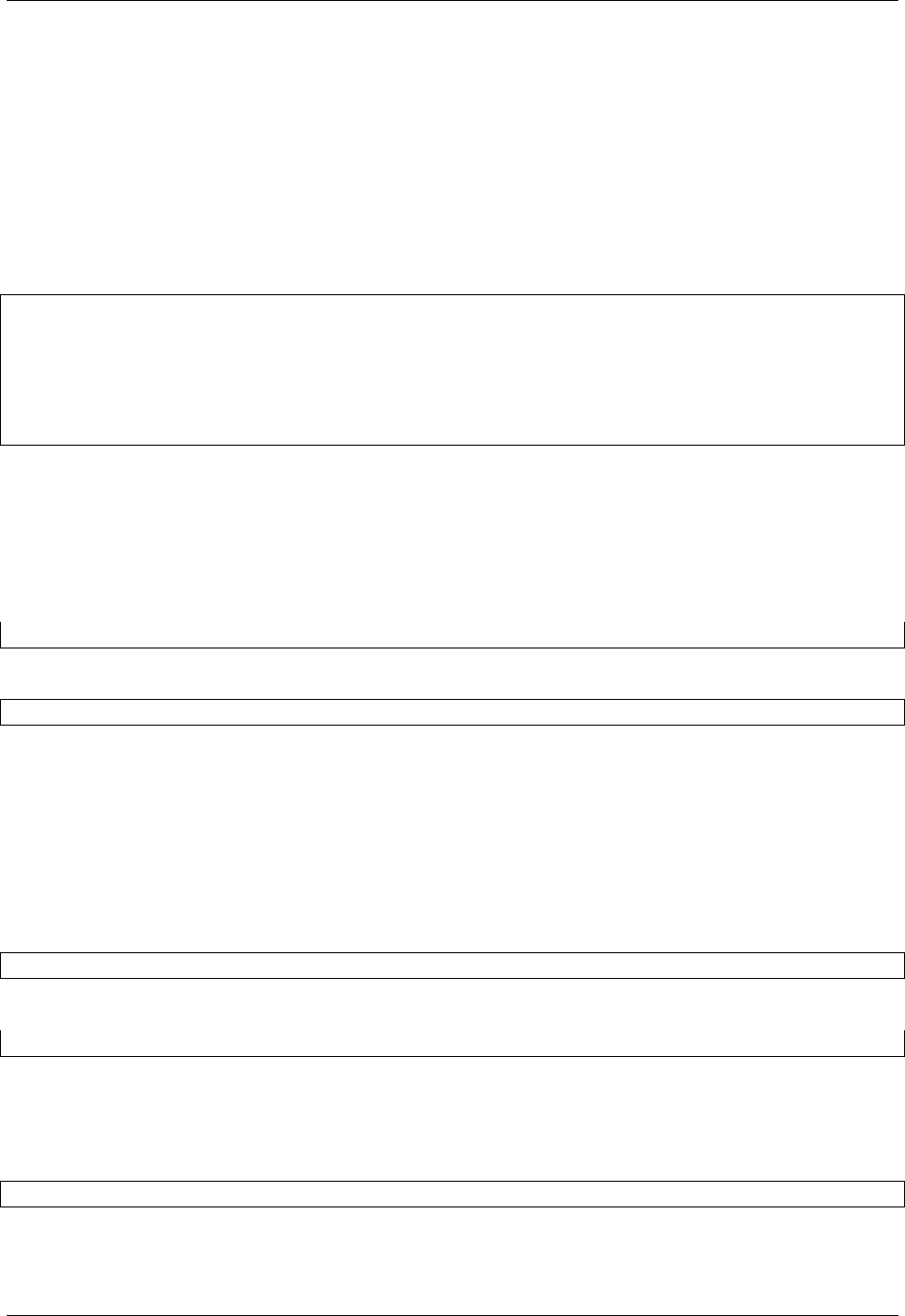
To convert data any pair of formats odo.odo relies on a network of pairwise conversions. We visualize that network
below
A single call to odo may traverse several intermediate formats calling on several conversion functions. These functions
are chosen because they are fast, often far faster than converting through a central serialization format.
2.1.3 URI strings
Odo uses strings refer to data outside of Python.
Some example uris include the following:
myfile.json
myfiles.
*
.csv'
postgresql://hostname::tablename
mongodb://hostname/db::collection
ssh://user@host:/path/to/myfile.csv
hdfs://user@host:/path/to/myfile.csv
6 Chapter 2. Contents
odo Documentation, Release 0.5.0+26.g55cec3c
Fig. 2.1: Each node represents a data format. Each directed edge represents a function to transform data between
two formats. A single call to odo may traverse multiple edges and multiple intermediate formats. Red nodes support
larger-than-memory data.
2.1. General 7

odo Documentation, Release 0.5.0+26.g55cec3c
What sorts of URI’s does odo support?
• Paths to files on disk
– .csv
– .json
– .txt/log
– .csv.gz/json.gz
– .hdf5
– .hdf5::/datapath
– .bcolz
– .xls(x)
– .sas7bdat
• Collections of files on disk
–
*
.csv
• SQLAlchemy strings
– sqlite:////absolute/path/to/myfile.db::tablename
– sqlite:////absolute/path/to/myfile.db (specify a particular table)
– postgresql://username:password@hostname:port
– impala://hostname (uses impyla)
– anything supported by SQLAlchemy
• MongoDB Connection strings
– mongodb://username:password@hostname:port/database_name::collection_name
• Remote locations via SSH, HDFS and Amazon’s S3
– ssh://user@hostname:/path/to/data
– hdfs://user@hostname:/path/to/data
– s3://path/to/data
Separating parts with ::
Many forms of data have two paths, the path to the file and then the path within the file. For example we refer to the
table accounts in a Postgres database like so:
postgresql://localhost::accounts
In this case the separator :: separates the database postgreqsl://localhost from the table within the
database, accounts.
This also occurs in HDF5 files which have an internal datapath:
myfile.hdf5::/path/to/data
8 Chapter 2. Contents

odo Documentation, Release 0.5.0+26.g55cec3c
Specifying protocols with ://
The database string sqlite:///data/my.db is specific to SQLAlchemy, but follows a common format, notably:
Protocol: sqlite://
Filename: data/my.db
Odo also uses protocols in many cases to give extra hints on how to handle your data. For example Python has a few
different libraries to handle HDF5 files (h5py, pytables, pandas.HDFStore). By default when we see a URI
like myfile.hdf5 we currently use h5py. To override this behavior you can specify a protocol string like:
hdfstore://myfile.hdf5
to specify that you want to use the special pandas.HDFStore format.
Note: sqlite strings are a little odd in that they use three slashes by default (e.g. sqlite:///my.db) and four
slashes when using absolute paths (e.g. sqlite:////Users/Alice/data/my.db).
How it works
We match URIs by to a collection of regular expressions. This is handled by the resource function.
>>> from odo import resource
>>> resource('sqlite:///data.db::iris')
Table('iris', MetaData(bind=Engine(sqlite:///myfile.db)), ...)
When we use a string in odo this is actually just shorthand for calling resource.
>>> from odo import odo
>>> odo('some-uri', list) # When you write this
>>> odo(resource('some-uri'), list) # actually this happens
Notably, URIs are just syntactic sugar, you don’t have to use them. You can always construct the object explic-
itly. Odo invents very few types, preferring instead to use standard projects within the Python ecosystem like
sqlalchemy.Table or pymongo.Collection. If your application also uses these types then it’s likely that
odo already works with your data.
Can I extend this to my own types?
Absolutely. Lets make a little resource function to load pickle files.
import pickle
from odo import resource
@resource.register('.
*
\.pkl') # match anything ending in .pkl
def resource_pickle(uri,
**
kwargs):
with open(uri) as f:
result = pickle.load(f)
return result
You can implement this kind of function for your own data type. Here we just loaded whatever the object was into
memory and returned it, a rather simplistic solution. Usually we return an object with a particular type that represents
that data well.
2.1. General 9
odo Documentation, Release 0.5.0+26.g55cec3c
2.1.4 Data Types
We can resolve errors and increase efficiency by explicitly specifying data types. Odo uses DataShape to specify
datatypes across all of the formats that it supports.
First we motivate the use of datatypes with two examples, then we talk about how to use DataShape.
Datatypes prevent errors
Consider the following CSV file:
name,balance
Alice,100
Bob,200
...
<many more lines>
...
Zelda,100.25
When odo loads this file into a new container (DataFrame, new SQL Table, etc.) it needs to know the datatypes of
the source so that it can create a matching target. If the CSV file is large then it looks only at the first few hundred
lines and guesses a datatype from that. In this case it might incorrectly guess that the balance column is of integer type
because it doesn’t see a decimal value until very late in the file with the line Zelda,100.25. This will cause odo
to create a target with the wrong datatypes which will foul up the transfer.
Odo will err unless we provide an explicit datatype. So we had this datashape:
var
*
{name: string, balance: int64)
But we want this one:
var
*
{name: string, balance: float64)
Datatypes increase efficiency
If we move that same CSV file into a binary store like HDF5 then we can significantly increase efficiency if we use
fixed-length strings rather than variable length. So we might choose to push all of the names into strings of length 100
instead of leaving their lengths variable. Even with the wasted space this is often more efficient. Good binary stores
can often compress away the added space but have trouble managing things of indeterminate length.
So we had this datashape:
var
*
{name: string, balance: float64}
But we want this one:
var
*
{name: string[100], balance: float64}
What is DataShape?
DataShape is a datatype system that includes scalar types:
string, int32, float64, datetime, ...
Option / missing value types:
10 Chapter 2. Contents

odo Documentation, Release 0.5.0+26.g55cec3c
?string, ?int32, ?float64, ?datetime, ...
Fixed length Collections:
10
*
int64
Variable length Collections:
var
*
int64
Record types:
{name: string, balance: float64}
And any composition of the above:
10
*
10
*
{x: int32, y: int32}
var
*
{name: string,
payments: var
*
{when: ?datetime, amount: float32}}
DataShape and odo
If you want to be explicit you can add a datashape to an odo call with the dshape= keyword
>>> odo('accounts.csv', pd.DataFrame,
... dshape='var
*
{name: string, balance: float64}')
This removes all of the guesswork from the odo heuristics. This can be necessary in tricky cases.
Use discover to get approximate datashapes
We rarely write out a full datashape by hand. Instead, use the discover function to get the datashape of an object.
>>> import numpy as np
>>> from odo import discover
>>> x = np.ones((5, 6), dtype='f4')
>>> discover(x)
dshape("5
*
6
*
float32")
In self describing formats like numpy arrays this datashape is guaranteed to be correct and will return very quickly. In
other cases like CSV files this datashape is only a guess and might need to be tweaked.
>>> from odo import odo, resource, discover
>>> csv = resource('accounts.csv') # Have to use resource to discover URIs
>>> discover(csv)
dshape("var
*
{name: string, balance: int64}")
>>> ds = dshape("var
*
{name: string, balance: float64") # copy-paste-modify
>>> odo('accounts.csv', pd.DataFrame, dshape=ds)
In these cases we can copy-paste the datashape and modify the parts that we need to change. In the example above we
couldn’t call discover directly on the URI, ’accounts.csv’, so instead we called resource on the URI first.
discover returns the datashape string on all strings, regardless of whether or not we intend them to be URIs.
2.1. General 11
odo Documentation, Release 0.5.0+26.g55cec3c
Learn More
DataShape is a separate project from odo. You can learn more about it at http://datashape.readthedocs.org/en/latest/
2.1.5 Drop
The odo.drop function deletes a data resource. That data resource may live outside of Python.
Examples
>>> from odo import drop
>>> drop('myfile.csv') # Removes file
>>> drop('sqlite:///my.db::accounts') # Drops table 'accounts'
>>> drop('myfile.hdf5::/data/path') # Deletes dataset from file
>>> drop('myfile.hdf5') # Deletes file
2.1.6 Loading CSVs into SQL Databases
When faced with the problem of loading a larger-than-RAM CSV into a SQL database from within Python, many
people will jump to pandas. The workflow goes something like this:
>>> import sqlalchemy as sa
>>> import pandas as pd
>>> con = sa.create_engine('postgresql://localhost/db')
>>> chunks = pd.read_csv('filename.csv', chunksize=100000)
>>> for chunk in chunks:
... chunk.to_sql(name='table', if_exist='append', con=con)
There is an unnecessary and very expensive amount of data conversion going on here. First we convert our CSV
into an iterator of DataFrames, then those DataFrames are converted into Python data structures compatible with
SQLAlchemy. Those Python objects then need to be serialized in a way that’s compatible with the database they are
being sent to. Before you know it, more time is spent converting data and serializing Python data structures than on
reading data from disk.
Use the technology that has already solved your problem well
Loading CSV files into databases is a solved problem. It’s a problem that has been solved well. Instead of rolling our
own loader every time we need to do this and wasting computational resources, we should use the native loaders in the
database of our choosing. Odo lets you do this with a single line of code.
How does odo achieve native database loading speed?
Odo uses the native CSV loading capabilities of the databases it supports. These loaders are extremely fast. Odo
will beat any other pure Python approach when loading large datasets. The following is a performance comparison
of loading the entire NYC taxi trip and fare combined dataset (about 33GB of text) into PostgreSQL, MySQL, and
SQLite3 using odo. Our baseline for comparison is pandas.
NB: I’m happy to hear about other optimizations that I may not be taking advantage of.
12 Chapter 2. Contents

odo Documentation, Release 0.5.0+26.g55cec3c
Timings
CSV → PostgreSQL (22m 64s)
• READS: ~50 MB/s
• WRITES: ~50 MB/s
The COPY command built into postgresql is quite fast. Odo generates code for the COPY command using a custom
SQLAlchemy expression.
In [1]: %time t = odo('all.csv', 'postgresql://localhost::nyc')
CPU times: user 1.43 s, sys: 330 ms, total: 1.76 s
Wall time: 22min 46s
PostgreSQL → CSV (21m 32s)
Getting data out of the database takes roughly the same amount of time as loading it in.
pg_bulkload Command Line Utility (13m 17s)
• READS: ~50 MB/s
• WRITES: ~50 MB/s
A special command line tool called pg_bulkload exists solely for the purpose of loading files into a postgresql
table. It achieves its speedups by disabling WAL (write ahead logging) and buffering. Odo doesn’t use this (yet)
because the installation requires several steps. There are also implications for data integrity when turning off WAL.
$ time ./pg_bulkload nyc2.ctl < all.csv
NOTICE: BULK LOAD START
NOTICE: BULK LOAD END
1 Rows skipped.
173179759 Rows successfully loaded.
0 Rows not loaded due to parse errors.
0 Rows not loaded due to duplicate errors.
0 Rows replaced with new rows.
./pg_bulkload nyc2.ctl < all.csv 26.14s user 33.31s system 7% cpu 13:17.31 total
CSV → MySQL (20m 49s)
In [1]: %time t = odo('all.csv', 'mysql+pymysql://localhost/test::nyc')
CPU times: user 1.32 s, sys: 304 ms, total: 1.63 s
Wall time: 20min 49s
• READS: ~30 MB/s
• WRITES: ~150 MB/s
MySQL → CSV (17m 47s)
In [1]: %time csv = odo('mysql+pymysql://localhost/test::nyc', 'nyc.csv')
CPU times: user 1.03 s, sys: 259 ms, total: 1.29 s
Wall time: 17min 47s
2.1. General 13

odo Documentation, Release 0.5.0+26.g55cec3c
• READS: ~30 MB/s
• WRITES: ~30 MB/s
Similar to PostgreSQL, MySQL takes roughly the same amount of time to write a CSV as it does to load it into a table.
CSV → SQLite3 (57m 31s*)
In [1]: dshape = discover(resource('all.csv'))
In [2]: %time t = odo('all.no.header.csv', 'sqlite:///db.db::nyc',
...: dshape=dshape)
CPU times: user 3.09 s, sys: 819 ms, total: 3.91 s
Wall time: 57min 31s
* Here, we call discover on a version of the dataset that has the header in the first line and we use a version of the
dataset without the header line in the sqlite3 .import command. This is sort of cheating, but I wanted to see what
the loading time of sqlite3’s import command was without the overhead of creating a new file without the header line.
SQLite3 → CSV (46m 43s)
• READS: ~15 MB/s
• WRITES: ~13 MB/s
In [1]: %time t = odo('sqlite:///db.db::nyc', 'nyc.csv')
CPU times: user 2.7 s, sys: 841 ms, total: 3.55 s
Wall time: 46min 43s
Pandas
• READS: ~60 MB/s
• WRITES: ~3-5 MB/s
I didn’t actually finish this timing because a single iteration of inserting 1,000,000 rows took about 4 minutes and there
would be 174 such iterations bringing the total loading time to:
.. code-block:: python
>>> 175
*
4 / 60.0
11.66...
11.66 hours!
Nearly 12 hours to insert 175 million rows into a postgresql database. The next slowest database (SQLite) is still 11x
faster than reading your CSV file into pandas and then sending that DataFrame to PostgreSQL with the to_pandas
method.
Final Thoughts
For getting CSV files into the major open source databases from within Python, nothing is faster than odo since it
takes advantage of the capabilities of the underlying database.
Don’t use pandas for loading CSV files into a database.
14 Chapter 2. Contents
odo Documentation, Release 0.5.0+26.g55cec3c
2.1.7 Adding a new Backend
Q: How do I add new nodes to the odo graph?
Extend Functions
We extend Odo by implementing a few functions for each new type
• discover - Return the DataShape of an object
• convert - Convert data to new type
• append - Append data on to existing data source
• resource - Identify data by a string URI
We extend each of these by writing new small functions that we decorate with types. Odo will then pick these up,
integrate them in to the network, and use them when appropriate.
Discover
Discover returns the DataShape of an object. Datashape is a potentially nested combination of shape and datatype. It
helps us to migrate metadata consistently as we migrate the data itself. This enables us to emerge with the right dtypes
even if we have to transform through potentially lossy formats.
Example
>>> discover([1, 2, 3])
dshape("3
*
int32")
>>> import numpy as np
>>> x = np.empty(shape=(3, 5), dtype=[('name', 'O'), ('balance', 'f8')])
>>> discover(x)
dshape("3
*
5
*
{name: string, balance: float64}")
Extend
We import discover from the datashape library and extend it with a type.
from datashape import discover, from_numpy
@discover(pd.DataFrame)
def discover_dataframe(df,
**
kwargs):
shape = (len(df),)
dtype = df.values.dtype
return from_numpy(shape, dtype)
In this simple example we rely on convenience functions within datashape to form a datashape from a numpy shape
and dtype. For more complex situations (e.g. databases) it may be necessary to construct datashapes manually.
Convert
Convert copies your data in to a new object with a different type.
2.1. General 15
odo Documentation, Release 0.5.0+26.g55cec3c
Example
>>> x = np.arange(5)
>>> x
array([0, 1, 2, 3, 4])
>>> convert(list, x)
[0, 1, 2, 3, 4]
>>> import pandas as pd
>>> convert(pd.Series, x)
0 0
1 1
2 2
3 3
4 4
dtype: int64
Extend
Import convert from odo and register it with two types, one for the target and one for the source
from odo import convert
@convert.register(list, np.ndarray)
def array_to_list(x,
**
kwargs):
return x.tolist()
@convert.register(pd.Series, np.ndarray)
def array_to_series(x,
**
kwargs):
return pd.Series(x)
Append
Append copies your data in to an existing dataset.
Example
>>> x = np.arange(5)
>>> x
array([0, 1, 2, 3, 4])
>>> L = [10, 20, 30]
>>> _ = append(L, x)
>>> L
[10, 20, 30, 0, 1, 2, 3, 4]
Extend
Import append from odo and register it with two types, one for the target and one for the source. Usually we teach
odo how to append from one preferred type and then use convert for all others
16 Chapter 2. Contents

odo Documentation, Release 0.5.0+26.g55cec3c
from odo import append
@append.register(list, list)
def append_list_to_list(tgt, src,
**
kwargs):
tgt.extend(src)
return tgt
@append.register(list, object) # anything else
def append_anything_to_list(tgt, src,
**
kwargs):
source_as_list = convert(list, src,
**
kwargs)
return append(tgt, source_as_list,
**
kwargs)
Resource
Resource creates objects from string URIs matched against regular expressions.
Example
>>> resource('myfile.hdf5')
<HDF5 file "myfile.hdf5" (mode r+)>
>>> resource('myfile.hdf5::/data', dshape='10
*
10
*
int32')
<HDF5 dataset "data": shape (10, 10), type "<i4">
The objects it returns are h5py.File and h5py.Dataset respectively. In the second case resource found that the
dataset did not exist so it created it.
Extend
We import resource from odo and register it with regular expressions
from odo import resource
import h5py
@resource.register('.
*
\.hdf5')
def resource(uri,
**
kwargs):
return h5py.File(uri)
General Notes
We pass all keyword arguments from the top-level call to odo to all functions. This allows special keyword arguments
to trickle down to the right place, e.g. delimiter=’;’ makes it to the pd.read_csv call when interacting with
CSV files, but also means that all functions that you write must expect and handle unwanted keyword arguments. This
often requires some filtering on your part.
Even though all four of our abstract functions have a .register method they operate in very different ways.
Convert is managed by networkx and path finding, append and discover are managed by multipledispatch, and
resource is managed by regular expressions.
Examples are useful. You may want to look at some of the odo source for simple backends for help
https://github.com/blaze/odo/tree/master/odo/backends
2.1. General 17

odo Documentation, Release 0.5.0+26.g55cec3c
2.1.8 Release Notes
Release 0.3.4
Release 0.3.4
Date September 15, 2015
New Features
• Added support for Spark 1.4 on Python 2.7 and Python 3.4 (#294)
Experimental Features
Warning: Experimental features are subject to change.
• Add support for specifying primary and foreign key relationships in the SQL backend (#274).
New Backends
None
Improved Backends
• Dialect discovery on CSV files now samples a subset of the file. This allows S3(CSV) to have correct values for
its dialect (#293).
• Loading a set of files on a s3 bucket with a prefix into a redshift database now works (#293).
API Changes
None
Bug Fixes
• Cope with Dask and bcolz API changes (#270).
• Fixed a bug where columns in dshape were being ignored when converting a numpy array to a DataFrame
(#273).
• Fix appending into a sql table from chunks not returning the table. (#278).
• Fix a bug where ’pytables://’ wasn’t being properly stripped off the URI (#292)
• Fix a bug where a non-existent header row was being removed from an S3(CSV) because the dialect was set
incorrectly (#293)
• Fix a bug where the SparkSQL backend wouldn’t work if we didn’t have paramiko installed (#300)
• Fix a testing bug where the endlines were being compared and they shouldn’t have been (#312).
• Fix a bug where sniffing multibyte encodings potentially chopped off part of the encoded string (#309, #311).
18 Chapter 2. Contents

odo Documentation, Release 0.5.0+26.g55cec3c
Miscellaneous
• Adds copydoc() function to copy docstrings from one object onto another. This helps with the pattern of
explicitly setting the __doc__ attribute to the __doc__ of another function or class. This function can be used
as a decorator like: @copydoc(FromThisClass) or as a function like: copydoc(FromThisClass,
to_this_function). (#277).
Release 0.3.3
Release 0.3.3
Date July 7th, 2015
New Backends
None
Improved Backends
• Implement SQL databases to CSV conversion using native the database dump (#174, #189, #191, #199).
• Improve CSV header existence inference (#192).
• Non-standard schemas can be passed into resource() with the schema argument (#223).
API Changes
• unicode strings can be passed in as URIs to resource() (#212).
Bug Fixes
• Fixed writing compressed CSVs in Python 3 and Windows (#188, #190).
• Dask API changes (#226).
• Fix some tests that would fail on binstar because they weren’t properly skipped (#216).
• PyTables API compatibility when given a integer valued float (#236).
• Default to None when plucking and a key isn’t found (#228).
• Fix gzip dispatching on JSON discovery (#243).
• ~odo.chunks.Chunks wrapping iterators can now be discovered without consuming the first element.
2.2 Formats
2.2.1 AWS
Dependencies
• boto
2.2. Formats 19

odo Documentation, Release 0.5.0+26.g55cec3c
• sqlalchemy
• psycopg2
• redshift_sqlalchemy
Setup
First, you’ll need some AWS credentials. Without these you can only access public S3 buckets. Once you have those,
S3 interaction will work. For other services such as Redshift, the setup is a bit more involved.
Once you have some AWS credentials, you’ll need to put those in a config file. Boto has a nice doc page on how to set
this up.
Now that you have a boto config, we’re ready to interact with AWS.
Interface
odo provides access to the following AWS services:
• S3 via boto.
• Redshift via a SQLAlchemy dialect
URIs
To access an S3 key, simply provide the path to the S3 key prefixed with s3://
>>> csvfile = resource('s3://bucket/key.csv')
S3 commonly uses a prefix to limit an operation to a subset of keys. We can simulate a glob of keys by combining
a prefix with the
*
character:
>>> csv_glob = resource('s3://bucket/prefix
*
.csv')
This will match all keys with starting with prefix and ending with the .csv extension. The result csv_glob can
be used just like a glob of files from your local disk.
Accessing a Redshift database is the same as accessing it through SQLAlchemy
>>> db = resource('redshift://user:pass@host:port/database')
To access an individual table simply append :: plus the table name
>>> table = resource('redshift://user:pass@host:port/database::table')
Conversions
odo can take advantage of Redshift’s fast S3 COPY command. It works transparently. For example, to upload a local
CSV file called users.csv to a Redshift table
>>> table = odo('users.csv', 'redshift://user:pass@host:port/db::users')
Remember that these are just additional nodes in the odo network, and as such, they are able to take advantage of
conversions to types that don’t have an explicit path defined for them. This allows us to do things like convert an S3
CSV to a pandas DataFrame
20 Chapter 2. Contents

odo Documentation, Release 0.5.0+26.g55cec3c
>>> import pandas as pd
>>> from odo import odo
>>> df = odo('s3://mybucket/myfile.csv', pd.DataFrame)
TODO
• Multipart uploads for huge files
• GZIP’d files
• JSON to Redshift (JSONLines would be easy)
• boto get_bucket hangs on Windows
2.2.2 CSV
Odo interacts with local CSV files through Pandas.
URIs
CSV URI’s are their paths/filenames
Simple examples of CSV uris:
myfile.csv
/path/to/myfile.csv.gz
Keyword Arguments
The standard csv dialect terms are usually supported:
has_header=True/False/None
encoding
delimiter
doublequote
escapechar
lineterminator
quotechar
quoting
skipinitialspace
However these or others may be in effect depending on what library is interacting with your file. Oftentimes this is the
pandas.read_csv function, which has an extensive list of keyword arguments
Conversions
The default paths in and out of CSV files is through Pandas DataFrames. Because CSV files might be quite large it
is dangerous to read them directly into a single DataFrame. Instead we convert them to a stream of medium sized
DataFrames. We call this type chunks(DataFrame).:
chunks(DataFrame) <-> CSV
CSVs can also be efficiently loaded into SQL Databases:
2.2. Formats 21

odo Documentation, Release 0.5.0+26.g55cec3c
CSV -> SQL
2.2.3 JSON
Odo interacts with local JSON files through the standard json library.
URIs
JSON URI’s are their paths/filenames
Simple examples of JSON uris:
myfile.json
/path/to/myfile.json.gz
Line Delimited JSON
Internally odo can deal with both traditional “single blob per file” JSON as well as line-delimited “one blob per line”
JSON. We inspect existing files to see which format it is. On new files we default to line-delimited however this can
be overruled by using the following protocols:
json://myfile.json # traditional JSON
jsonlines://myfile.json # line delimited JSON
Conversions
The default paths in and out of JSON files is through Python iterators of dicts.:
JSON <-> Iterator
2.2.4 HDF5
The Hierarchical Data Format is a binary, self-describing format, supporting regular strided and random access. There
are three main options in Python to interact with HDF5
• h5py - an unopinionated reflection of the HDF5 library
• pytables - an opinionated version, adding extra features and conventions
• pandas.HDFStore - a commonly used format among Pandas users.
All of these libraries create and read HDF5 files. Unfortunately some of them have special conventions that can only
be understood by their library. So a given HDF5 file created some of these libraries may not be well understood by the
others.
Protocols
If given an explicit object (not a string uri), like an h5py.Dataset, pytables.Table or pandas.HDFStore
then the odo project can intelligently decide what to do. If given a string, like myfile.hdf5::/data/path then
odo defaults to using the vanilla h5py solution, the least opinionated of the three.
You can specify that you want a particular format with one of the following protocols
22 Chapter 2. Contents
odo Documentation, Release 0.5.0+26.g55cec3c
• h5py://
• pytables://
• hdfstore://
Limitations
Each library has limitations.
• H5Py does not like datetimes
• PyTables does not like variable length strings,
• Pandas does not like non-tabular data (like ndarrays) and, if users don’t select the format=’table’
keyword argument, creates HDF5 files that are not well understood by other libraries.
Our support for PyTables is admittedly weak. We would love contributions here.
URIs
A URI to an HDF5 dataset includes a filename, and a datapath within that file. Optionally it can include a protocol
Examples of HDF5 uris:
myfile.hdf5::/data/path
hdfstore://myfile.h5::/data/path
Conversions
The default paths in and out of HDF5 files include sequences of Pandas DataFrames and sequences of NumPy
ndarrays.:
h5py.Dataset <-> chunks(np.ndarray)
tables.Table <-> chunks(pd.DataFrame)
pandas.AppendableFrameTable <-> chunks(pd.DataFrame)
pandas.FrameFixed <-> DataFrame
2.2.5 Hadoop File System
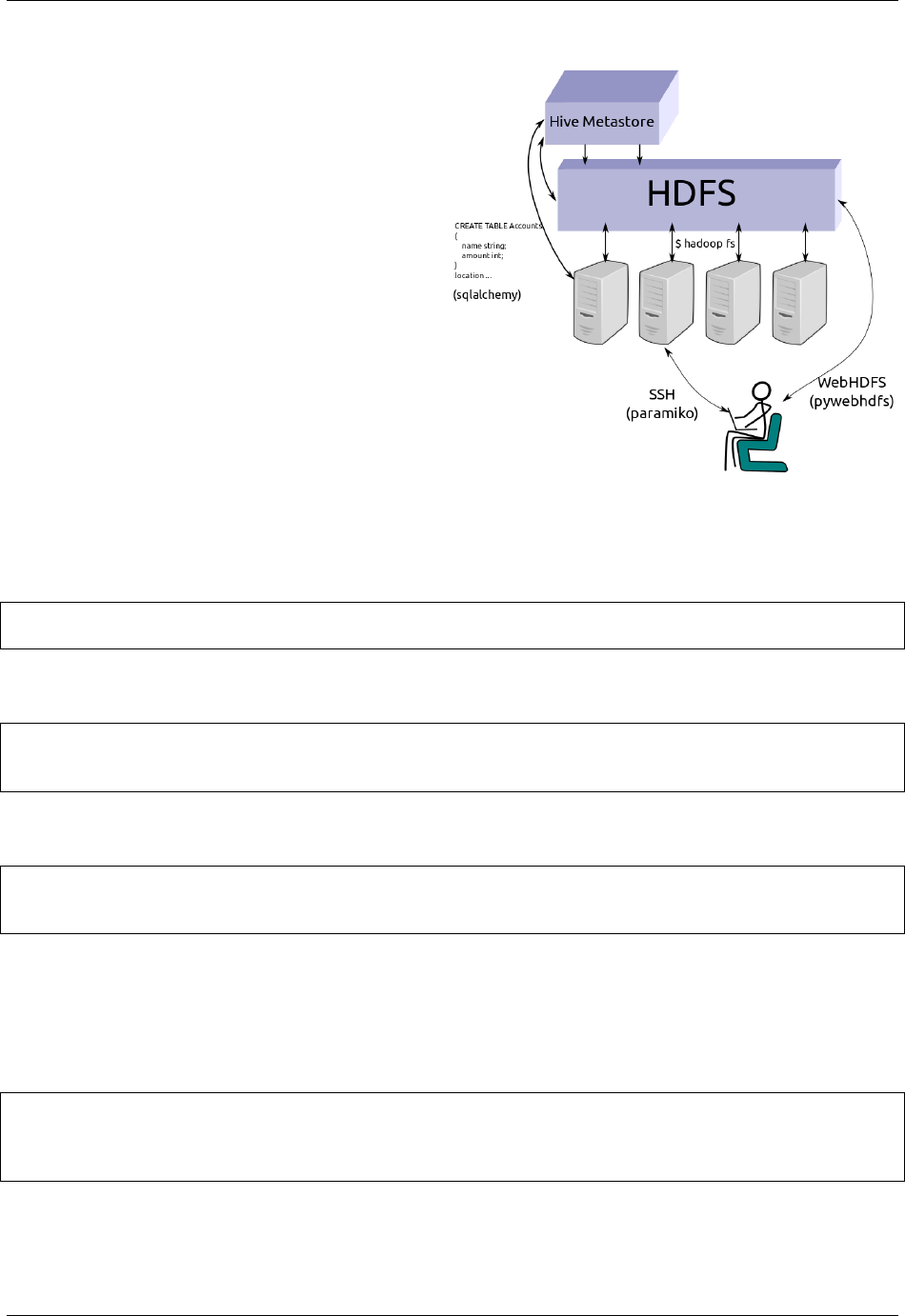
Odo interacts with the Hadoop File System using WebHDFS and the pywebhdfs Python lirary.
2.2. Formats 23
odo Documentation, Release 0.5.0+26.g55cec3c
URIs
HDFS uris consist of the hdfs:// protocol, a hostname, and a filename. Simple and complex examples follow:
hdfs://hostname:myfile.csv
hdfs://username@hostname:/path/to/myfile.csv
Alternatively you may want to pass authentication information through keyword arguments to the odo function as in
the following example
>>> from odo import odo
>>> odo('localfile.csv', 'hdfs://hostname:myfile.csv',
... port=14000, user='hdfs')
We pass through authentication keyword arguments to the pywebhdfs.webhdfs.PyWebHdfsClient class,
using the following defaults:
user_name='hdfs'
host=None
port='14000'
Constructing HDFS Objects explicitly
Most users usually interact with odo using URI strings.
Alternatively you can construct objects programmatically. HDFS uses the HDFS type modifier
>>> auth = {'user': 'hdfs', 'port': 14000, 'host': 'hostname'}
>>> data = HDFS(CSV)('/user/hdfs/data/accounts.csv',
**
auth)
>>> data = HDFS(JSONLines)('/user/hdfs/data/accounts.json',
**
auth)
>>> data = HDFS(Directory(CSV))('/user/hdfs/data/',
**
auth)
24 Chapter 2. Contents

odo Documentation, Release 0.5.0+26.g55cec3c
Conversions
We can convert any text type (CSV, JSON, JSONLines, TextFile) to its equivalent on HDFS (HDFS(CSV),
HDFS(JSON), ...). The odo network allows conversions from other types, like a pandas dataframe to a CSV file
on HDFS, by routing through a temporary local csv file.:
HDFS(
*
) <->
*
Additionally we know how to load HDFS files into the Hive metastore:
HDFS(Directory(CSV)) -> Hive
The network also allows conversions from other types, like a pandas DataFrame to an HDFS CSV file, by routing
through a temporary local csv file.:
Foo <-> Temp(
*
) <-> HDFS(
*
)
2.2.6 Hive Metastore
The Hive metastore relates SQL metadata to files on the Hadoop File System (HDFS). It is similar to a SQL database
in that it contains information about SQL tables but dissimilar in that data isn’t stored in Hive but remains ordinary
files on HDFS.
Odo interacts with Hive mostly through sqlalchemy and also with a bit of custom code due to its peculiarities.
URIs
Hive uris match exactly SQLAlchemy connection strings with the hive:// protocol. Additionally, Impala, another
SQL database on HDFS can also connect to the same tables.
hive://hostname
hive://user@hostname:port/database-name
hive://user@hostname:port/database-name::table-name
impala://hostname::table-name
Additionally you should probably inspect docs on HDFS due to the tight integration between the two.
Options
Hive tables have a few non-standard options on top of normal SQL:
stored_as - File format on disk like TEXTFILE, PARQUET, ORC
path - Absolute path to file location on HDFS
external=True - Whether to keep the file external or move it to Hive
directory
See Hive DDL docs for more information.
Conversions
We commonly load CSV files in to Hive, either from HDFS or from local disk on one of the machines that comprise
the HDFS cluster:
2.2. Formats 25

odo Documentation, Release 0.5.0+26.g55cec3c
HDFS(Directory(CSV)) -> Hive
SSH(Directory(CSV)) -> Hive
SSH(CSV) -> Hive
Additionally we can use Hive to efficiently migrate this data to new data in a different format:
Hive -> Hive
And as with all SQL systems through SQLAlchemy we can convert a Hive table to a Python Iterator, though this is
somewhat slow:
Hive -> Iterator
Impala
Impala operates on the same data as Hive, is generally faster, though also has a couple of quirks.
While Impala connects to the same metastore it must connect to one of the worker nodes, not the same head node to
which Hive connects. After you load data in to hive you need to send the invalidate metadata to Impala.
>>> odo('hdfs://hostname::/path/to/data/
*
.csv', 'hive://hostname::table')
>>> imp = resource('impala://workernode')
>>> imp.connect().execute('invalidate metadata')
This is arguably something that odo should handle in the future. After this, all tables in Hive are also available to
Impala.
You may want to transform your data in to Parquet format for efficient querying. A two minute query on Hive in CSV
might take one minute on Hive in Parquet and only three seconds in Impala in Parquet.
>>> odo('hive://hostname::table', 'hive://hostname::table_parquet',
... external=False, stored_as='PARQUET')
2.2.7 Mongo
Odo interacts with Mongo databases through PyMongo.
URIs
Simple and complex examples of MongoDB uris:
mongodb://localhost/mydb::mycollection
mongodb://user:password@localhost:port/mydb::mycollection
Conversions
The default path in and out of a Mongo database is to use the PyMongo library to produce and consume iterators of
Python dictionaries.:
pymongo.Collection <-> Iterator
26 Chapter 2. Contents

odo Documentation, Release 0.5.0+26.g55cec3c
2.2.8 Spark/SparkSQL
Dependencies
• spark
• pyhive
• sqlalchemy
Setup
We recommend you install Spark via conda from the blaze binstar channel:
$ conda install pyhive spark -c blaze
The package works well on Ubuntu Linux and Mac OS X. Other issues may arise when installing this package on a
non-Ubuntu Linux distro. There’s a known issue with Arch Linux.
Interface
Spark diverges a bit from other areas of odo due to the way it works. With Spark, all objects are attached to a special
object called SparkContext. There can only be one of these running at a time. In contrast, SparkSQL objects all
live inside of one or more SQLContext objects. SQLContext objects must be attached to a SparkContext.
Here’s an example of how to setup a SparkContext:
>>> from pyspark import SparkContext
>>> sc = SparkContext('app', 'local')
Next we create a SQLContext:
>>> from pyspark.sql import SQLContext
>>> sql = SQLContext(sc) # from the previous code block
From here, you can start using odo to create SchemaRDD objects, which are the SparkSQL version of a table:
>>> from odo import odo
>>> data = [('Alice', 300.0), ('Bob', 200.0), ('Donatello', -100.0)]
>>> type(sql)
<class 'pyspark.sql.SQLContext'>
>>> srdd = odo(data, sql, dshape='var
*
{name: string, amount: float64}')
>>> type(srdd)
<class 'pyspark.sql.SchemaRDD'>
Note the type of srdd. Usually odo(A, B) will return an instance of B if B is a type. With Spark and SparkSQL,
we need to attach whatever we make to a context, so we “append” to an existing SparkContext/SQLContext.
Instead of returning the context object, odo will return the SchemaRDD that we just created. This makes it more
convenient to do things with the result.
This functionality is nascent, so try it out and don’t hesitate to report a bug or request a feature!
URIs
URI syntax isn’t currently implemented for Spark objects.
2.2. Formats 27

odo Documentation, Release 0.5.0+26.g55cec3c
Conversions
The main paths into and out of RDD and SchemaRDD are through Python list objects:
RDD <-> list
SchemaRDD <-> list
Additionally, there’s a specialized one-way path for going directly to SchemaRDD from RDD:
RDD -> SchemaRDD
TODO
• Resource/URIs
• Native loaders for JSON and possibly CSV
• HDFS integration
2.2.9 SAS
Odo interacts with local sas7bdat files through sas7bdat.
URIs
SAS URI’s are their paths/filenames
Simple examples of SAS uris:
myfile.sas7bdat
/path/to/myfile.sas7bdat
Conversions
The default paths out of SAS files is through Python iterators and Pandas DataFrames.
SAS7BDAT -> Iterator SAS7BDAT -> pd.DataFrame
This is a closed file format with nice but incomplete support from the sas7bdat Python library. You should not expect
comprehensive coverage.
2.2.10 SQL
Odo interacts with SQL databases through SQLAlchemy. As a result, odo supports all databases that SQLAlchemy
supports. Through third-party extensions, SQLAlchemy supports most databases.
28 Chapter 2. Contents

odo Documentation, Release 0.5.0+26.g55cec3c
Warning: When putting an array-like object such as a NumPy array into a database you must provide the column
names in the form of a Record datashape.
Note: Without column names, it doesn’t make sense to put an array into a database table, since a database table
doesn’t make sense without named columns. The inability to put an array with unnamed columns into a database
is intentional.
Here’s a failing example:
>>> import numpy as np
>>> from odo import odo
>>> x = np.zeros((10, 2))
>>> t = odo(x, 'sqlite:///db.db::x') # this will NOT work
Traceback (most recent call last):
...
TypeError: dshape measure must be a record type e.g., "{a: int64, b: int64}". Input measure is ctype("float64")
Here’s what to do instead:
>>> t = odo(x, 'sqlite:///db.db::x', # works because columns are named
>>> ... dshape='var
*
{a: float64, b: float64}')
URIs
Simple and complex examples of SQL uris:
postgresql://localhost::accounts
postgresql://username:[email protected]:10000/default::accounts
SQL uris consist of the following
• dialect protocol: postgresql://
• Optional authentication information: username:password@
• A hostname or network location with optional port: 54.252.14.53:10000
• Optional database/schema name: /default
• A table name with the :: separator: ::accounts
Executing Odo Against Databases
Sqlalchemy allows objects to be bound to a particular database connection. This is known as the ‘bind’ of the object,
or that the object is ‘bound’.
By default, odo expects to be working with either bound sqlalchemy objects or uris to tables.
For example, when working with a sqlalchemy object, one must be sure to pass a bound metadata to the construction
of your tables.
>>> import sqlalchemy as sa
>>> sa.MetaData()
>>> tbl = sa.Table(
... 'tbl',
... metadata,
... sa.Column('a', sa.Integer, primary_key=True),
2.2. Formats 29

odo Documentation, Release 0.5.0+26.g55cec3c
... )
>>> odo([[1], [2], [3]], tbl, dshape='var
*
{a: int}') # this will NOT work
Traceback (most recent call last):
...
UnboundExecutionError: Table object 'tbl' is not bound to an Engine or Connection. Execution can not proceed without a database to execute against.
We have two options for binding metadata to objects, we can explicitly bind our tables, or we can pass it to odo as a
keyword argument.
Here is an example of constructing the table with a bound metadata:
>>> import sqlalchemy as sa
>>> metadata = sa.MetaData(bind='sqlite:///db.db') # NOTE: pass the uri to the db here
>>> tbl = sa.Table(
... 'tbl',
... metadata,
... sa.Column('a', sa.Integer, primary_key=True),
... )
>>> odo([[1], [2], [3]], tbl) # this know knows where to field the table.
Here is an example of passing the bind to odo:
>>> import sqlalchemy as sa
>>> sa.MetaData()
>>> tbl = sa.Table(
... 'tbl',
... metadata,
... sa.Column('a', sa.Integer, primary_key=True),
... )
>>> bind = 'sqlite:///db.db'
>>> odo([[1], [2], [3]], tbl, dshape='var
*
{a: int}', bind=bind) # pass the bind to odo here
Here, the bind may be either a uri to a database, or a sqlalchemy Engine object.
Conversions
The default path in and out of a SQL database is to use the SQLAlchemy library to consume iterators of Python
dictionaries. This method is robust but slow.:
sqlalchemy.Table <-> Iterator
sqlalchemy.Select <-> Iterator
For a growing subset of databases (sqlite, MySQL, PostgreSQL, Hive, Redshift) we also use the CSV or JSON tools
that come with those databases. These are often an order of magnitude faster than the Python->SQLAlchemy route
when they are available.:
sqlalchemy.Table <- CSV
Primary and Foreign Key Relationships
New in version 0.3.4.
Warning: Primary and foreign key relationship handling is an experimental feature and is subject to change.
Odo has experimental support for creating and discovering relational database tables with primary keys and foreign
key relationships.
30 Chapter 2. Contents
odo Documentation, Release 0.5.0+26.g55cec3c
Creating a new resource with a primary key
We create a new sqlalchemy.Table object with the resource function, specifying the primary key in the
primary_key argument
>>> from odo import resource
>>> dshape = 'var
*
{id: int64, name: string}'
>>> products = resource(
... 'sqlite:///db.db::products',
... dshape=dshape,
... primary_key=['id'],
... )
>>> products.c.id.primary_key
True
Compound primary keys are created by passing the list of columns that form the primary key. For example
>>> dshape = """
... var
*
{
... product_no: int32,
... product_sku: string,
... name: ?string,
... price: ?float64
... }
... """
>>> products = resource(
... 'sqlite:///%s::products' % fn,
... dshape=dshape,
... primary_key=['product_no', 'product_sku']
... )
Here, the column pair product_no, product_sku make up the compound primary key of the products table.
Creating resources with foreign key relationships
Creating a new resource with a foreign key relationship is only slightly more complex.
As a motivating example, consider two tables products and orders. The products table will be the table from
the primary key example. The orders table will have a many-to-one relationship to the products table. We can
create this like so
>>> orders_dshape = """
... var
*
{
... order_id: int64,
... product_id: map[int64, {id: int64, name: string}]
... }
... """
>>> orders = resource(
... 'sqlite:///db.db::orders',
... dshape=orders_dshape,
... primary_key=['order_id'],
... foreign_keys={
... 'product_id': products.c.id,
... }
... )
>>> products.c.id in orders.c.product_id.foreign_keys
True
2.2. Formats 31

odo Documentation, Release 0.5.0+26.g55cec3c
There are two important things to note here.
1. The general syntax for specifying the type of referring column is
map[<referring column type>, <measure of the table being referred to>]
2. Knowing the type isn’t enough to specify a foreign key relationship. We also need to know the table that has the
columns we want to refer to. The foreign_keys argument to the resource() function fills this need. It accepts
a dictionary mapping referring column names to referred to sqlalchemy.Column instances or strings such
as products.id.
There’s also a shortcut syntax using type variables for specifying foreign key relationships whose referred-to tables
have very complex datashapes.
Instead of writing our orders table above as
var
*
{order_id: int64, product_id: map[int64, {id: int64, name: string}]}
We can replace the value part of the map type with any word starting with a capital letter. Often this is a single capital
letter, such as T
var
*
{order_id: int64, product_id: map[int64, T]}
Odo will automatically fill in the datashape for T by calling discover() on the columns passed into the foreign_keys
keyword argument.
Finally, note that discovery of primary and foreign keys is done automatically if the relationships already exist in the
database so it isn’t necessary to specify them if they’ve already been created elsewhere.
More Complex Foreign Key Relationships
Odo supports creation and discovery of self referential foreign key relationships as well as foreign keys that are
elements of a compound primary key. The latter are usually seen when creating a many-to-many relationship via a
junction table.
Self referential relationships are most easily specified using type variables (see the previous section for a description
of how that works). Using the example of a management hierarchy:
>>> dshape = 'var
*
{eid: int64, name: ?string, mgr_eid: map[int64, T]}'
>>> t = resource(
... 'sqlite:///%s::employees' % fn,
... dshape=dshape,
... primary_key=['eid'],
... foreign_keys={'mgr_eid': 'employees.eid'}
... )
Note: Currently odo only recurses one level before terminating as we don’t yet have a syntax for truly
expressing recursive types in datashape
Here’s an example of creating a junction table (whose foreign keys form a compound primary key) using a modified
version of the traditional suppliers and parts database:
>>> suppliers = resource(
... 'sqlite:///%s::suppliers' % fn,
... dshape='var
*
{id: int64, name: string}',
... primary_key=['id']
... )
>>> parts = resource(
32 Chapter 2. Contents

odo Documentation, Release 0.5.0+26.g55cec3c
... 'sqlite:///%s::parts' % fn,
... dshape='var
*
{id: int64, name: string, region: string}',
... primary_key=['id']
... )
>>> suppart = resource(
... 'sqlite:///%s::suppart' % fn,
... dshape='var
*
{supp_id: map[int64, T], part_id: map[int64, U]}',
... primary_key=['supp_id', 'part_id'],
... foreign_keys={
... 'supp_id': suppliers.c.id,
... 'part_id': parts.c.id
... }
... )
>>> from odo import discover
>>> print(discover(suppart))
var
*
{
supp_id: map[int64, {id: int64, name: string}],
part_id: map[int64, {id: int64, name: string, region: string}]
}
Foreign Key Relationship Failure Modes
Some databases support the notion of having a foreign key reference one column from another table’s compound
primary key. For example
>>> product_dshape = """
... var
*
{
... product_no: int32,
... product_sku: string,
... name: ?string,
... price: ?float64
... }
... """
>>> products = resource(
... 'sqlite:///%s::products' % fn,
... dshape=product_dshape,
... primary_key=['product_no', 'product_sku']
... )
>>> orders_dshape = """
... var
*
{
... order_id: int32,
... product_no: map[int32, T],
... quantity: ?int32
... }
... """
>>> orders = resource(
... 'sqlite:///%s::orders' % fn,
... dshape=orders_dshape,
... primary_key=['order_id'],
... foreign_keys={
... 'product_no': products.c.product_no
... # no reference to product_sku, okay for sqlite, but not postgres
... }
... )
Here we see that when the orders table is constructed, only one of the columns contained in the primary key of the
products table is included.
2.2. Formats 33

odo Documentation, Release 0.5.0+26.g55cec3c
SQLite is an example of one database that allows this. Other databases such as PostgreSQL will raise an error if the
table containing the foreign keys doesn’t have a reference to all of the columns of the compound primary key.
Odo has no opinion on this, so if the database allows it, then odo will allow it. This is an intentional choice.
However, it can also lead to confusing situations where something works with SQLite, but not with PostgreSQL. These
are not bugs in odo, they are an explicit choice to allow flexibility with potentially large already-existing systems.
Amazon Redshift
When using Amazon Redshift the error reporting leaves much to be desired. Many errors look like this:
InternalError: (psycopg2.InternalError) Load into table 'tmp0' failed. Check 'stl_load_errors' system table for details.
If you’re reading in CSV data from S3, check to make sure that
1. The delimiter is correct. We can’t correctly infer everything, so you may have to pass that value in as e.g.,
delimiter=’|’.
2. You passed in the compression=’gzip’ keyword argument if your data are compressed as gzip files.
If you’re still getting an error and you’re sure both of the above are correct, please report a bug on the odo issue tracker
We have an open issue (#298) to discuss how to better handle the problem of error reporting when using Redshift.
2.2.11 SSH
Odo interacts with remote data over ssh using the paramiko library.
URIs
SSH uris consist of the ssh:// protocol, a hostname, and a filename. Simple and complex examples follow:
ssh://hostname:myfile.csv
ssh://username@hostname:/path/to/myfile.csv
Additionally you may want to pass authentication information through keyword arguments to the odo function as in
the following example
>>> from odo import odo
>>> odo('localfile.csv', 'ssh://hostname:myfile.csv',
... username='user', key_filename='.ssh/id_rsa', port=22)
We pass through authentication keyword arguments to the paramiko.SSHClient.connect method. That
method takes the following options:
port=22
username=None
password=None
pkey=None
key_filename=None
timeout=None
allow_agent=True
look_for_keys=True
compress=False
sock=None
34 Chapter 2. Contents

odo Documentation, Release 0.5.0+26.g55cec3c
Constructing SSH Objects explicitly
Most users usually interact with odo using URI strings.
Alternatively you can construct objects programmatically. SSH uses the SSH type modifier
>>> from odo import SSH, CSV, JSON
>>> auth = {'user': 'ubuntu',
... 'host': 'hostname',
... 'key_filename': '.ssh/id_rsa'}
>>> data = SSH(CSV)('data/accounts.csv',
**
auth)
>>> data = SSH(JSONLines)('accounts.json',
**
auth)
Conversions
We’re able to convert any text type (CSV, JSON, JSONLines, TextFile) to its equivalent on the remote
server (SSH(CSV), SSH(JSON), ...).:
SSH(
*
) <->
*
The network also allows conversions from other types, like a pandas DataFrame to a remote CSV file, by routing
through a temporary local csv file.:
Foo <-> Temp(
*
) <-> SSH(
*
)
2.3 Developer Documentation
2.3.1 Type Modifiers
Odo decides what conversion functions to run based on the type (e.g. pd.DataFrame, sqlalchemy.Table,
odo.CSV of the input. In many cases we want slight variations to signify different circumstances such as the differ-
ence between the following CSV files
• A local CSV file
• A sequence of CSV files
• A CSV file on a remote machine
• A CSV file on HDFS
• A CSV file on S3
• A temporary CSV file that should be deleted when we’re done
In principle we need to create subclasses for each of these and for their JSON, TextFile, etc. equivalents. To assist
with this we create functions to create these subclasses for us. These functions are named the following:
chunks - a sequence of data in chunks
SSH - data living on a remote machine
HDFS - data living on Hadoop File system
S3 - data living on Amazon's S3
Directory - a directory of data
Temp - a temporary piece of data to be garbage collected
We use these functions on types to construct new types.
2.3. Developer Documentation 35
odo Documentation, Release 0.5.0+26.g55cec3c
>>> SSH(CSV)('/path/to/data', delimiter=',', user='ubuntu')
>>> Directory(JSON)('/path/to/data/')
We compose these functions to specify more complex situations like a temporary directory of JSON data living on S3
>>> Temp(S3(Directory(JSONLines)))
Use URIs
Most users don’t interact with these types. They are for internal use by developers to specify the situations in which a
function should be called.
chunks
A particularly important type modifier is chunks, which signifies an iterable of some other type. For example
chunks(list) means an iterable of Python lists and chunks(pd.DataFrame) an iterable of DataFrames. The
chunks modifier is often used to convert between two out-of-core formats via an in-core format. This is also a nice
mechanism to interact with data in an online fashion
>>> from odo import odo, chunks
>>> import pandas as pd
>>> seq = odo('postgresql://localhost::mytable', chunks(pd.DataFrame))
>>> for df in seq:
... # work on each dataframe sequentially
chunks may also be used to write an iterable of chunks into another resource. For example, we may use chunks to
write a sequence of numpy arrays into a postgres table while only ever holding one whole array in memory like so:
>>> import numpy as np
>>> from odo import odo, chunks
>>> seq = (np.random.randn(5, 3) for _ in range(3))
>>> odo(chunks(np.ndarray)(seq), 'postgresql://localhost::mytable')
chunks(type_)(seq) is merely a small box wrapping the inner sequence that allows odo to know the types of
the elements in the sequence. We may still use this sequence as we would any other, including looping over it.
Because this is wrapping the inner sequence, we may only iterate over the chunks multiple times if the inner sequence
supports being iterated over more than once. For example:
>>> from odo import chunks
>>> CL = chunks(list)
>>> multuple_iteration_seq = CL([[0, 1, 2], [3, 4, 5])
>>> tuple(multuple_iteration_seq)
([0, 1, 2], [3, 4, 5])
>>> tuple(multuple_iteration_seq)
([0, 1, 2], [3, 4, 5])
>>> single_iteration_seq = CL(iter([[0, 1, 2], [3, 4, 5]]))
>>> tuple(single_iteraton_seq)
([0, 1, 2], [3, 4, 5])
>>> tuple(single_iteration_seq)
()
36 Chapter 2. Contents

odo Documentation, Release 0.5.0+26.g55cec3c
2.3.2 Four Operations
The Blaze project originally included odo.odo as a magic function that moved data between containers. This func-
tion was both sufficiently useful and sufficiently magical that it was moved to a separate project, its functionality
separated into three operations
1. convert: Transform dataset to a new type. convert(list, (1, 2, 3))
2. append: Append a dataset to another. append([], (1, 2, 3))
3. resource: Obtain or create dataset from a URI string resource(’myfile.csv’)
These are magically tied together as the original odo function
4. odo: Put stuff into other stuff (deliberately vague/magical.)
Odo is part of the Open Source Blaze projects supported by Continuum Analytics
2.3. Developer Documentation 37

